By David Kale and Zachary Lipton
Starting Friday, August 18th and lasting two days, Northeastern University in Boston hosted the eighth annual Machine Learning for Healthcare (MLHC) conference. This year marked MLHC’s second year as a publishing conference with an archival proceedings in the Journal of Machine Learning Research (JMLR). Incidentally, the transition to formal publishing venue in 2016 coincided with the name change to MLHC from Meaningful Use of Complex Medical Data, denoted by the memorable acronym MUCMD (pronounced MUCK-MED).
From its beginnings at Children’s Hospital Los Angeles as a non-archival symposium, the meeting set out to address the following problem:
- Machine learning, even then, was seen as a powerful tool that can confer insights and improve processes in domains with well-defined problems and large quantities of interesting data.
- In the course of treating patients, hospitals produce massive streams of data, including vital signs, lab tests, medication orders, radiologic imaging, and clinical notes, and record many health outcomes of interest, e.g., diagnoses. Moreover, numerous tasks in clinical care present as well-posed machine learning problems.
- However, despite the clear opportunities, there was surprisingly little collaboration between machine learning experts and clinicians. Few papers at elite machine learning conferences addressed problems in clinical health and few machine learning papers were submitted to the elite medical journals.
The lack of work at the intersection of ML and healthcare may owe partly to the intrinsic difficulty of the problems. But many of us in this small intersection believe the lack of progress owes in large part to less glamourous causes, principally the bureaucratic overhead of acquiring data and the social overhead of forging interdisciplinary collaborations.
Consider the workflow for a typical machine learning research project. Step 1: Researcher has idea, thinks it might be useful for computer vision. Step 2: a researcher might go out and download a large public dataset like ImageNet over several hours. Step 3: Researcher gets down to business and does the actual technical work. Note that in this pipeline, getting to the hacking in Step 3 often takes only a handful of hours.
In contrast, acquiring medical data requires a deeper investment of energy. A project might first require befriending and building trust with clinicians and convincing them of machine learning’s potential to impact their problems. Even then, acquiring data requires negotiating appropriate data use agreements, preparing appropriately secure computer systems, and extracting and anonymizing data from the electronic health records (EHRs) and other hospital computer systems. From there, the machine learner must invest further time understanding, cleaning, and preprocessing the data.
But while these latter problems are headaches, like renewing a passport, they’re solvable with enough elbow grease. On the other hand, identifying the right problems and forging the initial collaborations requires considerably more imagination. The original MUCMD symposia were founded with the idea forging such collaborations. Doctors came pitching problems, machine learners came pitching methods. Now, in 2017, some of these collaborations have come to fruition, and MLHC has grown to provide a venue for this work. In the rest of this post, we’ll relay our takeaways from the invited talks, highlight research trends among the accepted papers, and discuss the form of the conference itself.

Following the conference, David Sontag’s student Irene Chen beat us to the punch with a succinct set of 30 takeaways. In this post, what we miss in timeliness, we make up for with volume. In this Pedant’s Guide to MLHC 2017, we present a full analysis of each talk, even the ones we missed.
Conflicts: (Dave Kale) sits among MLHC’s founding organizers, both of us have been involved as both reviewers and authors, and our own long-standing collaboration was born of a chance meeting at MUCMD in 2013.
Program
The MLHC 2017 program spanned two days, broken up into four half-day sessions and one poster session on the first evening. The single-track program included eight invited speakers, spotlight talks for all accepted papers and abstracts, an interdisciplinary panel, and ample time for coffee and discussion. While this year’s attendees skewed to the machine learning side (a demographic shift we hope to address in 2018), the invited speakers represented clinicians, health system administrators, machine learning scientists, and health industry startups. If you’d rather watch the talks for yourself than read our takeaways, you can find unedited videos of the sessions on the conference Facebook Event (individual talk videos will be available at a later date).
Friday, August 18th
John Halamka – CIO of Beth Israel Medical Center
Following a brief welcome from the MLHC organizers, the conference began with a call to action by John Halamka, CIO of Beth Israel Deaconess Medical Center (BIDMC) and Harvard Medical School, and apparently an early-rising organic farmer. Compared to the average technical talk, Halamka’s was unusually polished. Overall, the differences in aesthetics, in rehearsal, and stage command across technical, medical, corporate and public sector talks was an experience worth noting.

Halamka’s talk was more inspirational than technical, and he used the opportunity to enumerate roughly a dozen opportunities where he envisioned using machine learning in healthcare. Applications spanned the procedural, e.g., automated recognition of scanned patient consent forms; the administrative, e.g., early identification of potentially poor clinic service based on negative sentiment in social media; and the clinical, e.g., personalized medicine. BIDMC has developed and deployed solutions for several of these problems.
Notably, John said that BIDMC uses cloud storage and runs machine learning-as-service tools provided by both Amazon and Google. When asked about privacy and security concerns, John explained that academics attempting to circumvent on-premise security policies actually poses a greater risk to data security. He suggested that delegating storage and security to a large cloud vendor reduces this risk because large vendors have the resources to enforce strong security policies while also enabling the access required by researchers.
Daniel Golden – FDA-Approved Deep Learning
The second speaker was Daniel Golden, Director of Machine Learning at Arterys, a startup that recently obtained first-of-its-kind FDA clearance for its deep learning-based cardiac MRI decision support software. Golden’s spoke mostly about Arterys’ DeepVentricle model, a fully convolutional network based on UNet that detects and segments the contours of the right ventricle endocardium and left ventricle epicardium and endocardium. Segmentation is the first step in estimating cardiac measures like stroke volume and tends to be a significant time sink for radiologists.
Golden detailed several of the unique challenges presented by clinical images, in particular the paucity of complete annotations: fewer than 20% of images in Arterys’ training data had all three contours annotated. Arterys’ solution involved training on the full but partially labeled data set and back-propagating the error signal for the available labels only. This yielded a more accurate model that produced cleaner segmentations. Golden also spoke about production deployment and regulatory compliance. In particular, he emphasized that Arterys’ focus is on helping clinicians do their jobs more efficiently, rather than replacing them.

One curious fact was the dearth of imaging papers in MLHC’s proceedings, despite the flurry of startup activity around radiology. This may be explained by the existence of conferences like MICCAI (Medical Image Computing and Computer Assisted Intervention), which has long been receptive to machine learning-based approaches. It may also indicate the maturity of the field and its focus on translation and commercialization, rather than basic research.
NOTE: the DeepVentricle paper is difficult to find, but Arterys recently published an updated model called FastVentricle based on the ENet architecture.
Regina Barzilay – How Can NLP Help Cure Cancer?
Friday’s afternoon session began with a personal story from Regina Barzilay of MIT about her her diagnosis with and subsequent treatment for breast cancer. While her treatment was ultimately successful, Barzilay found herself dismayed by the state of clinical decision support. She (perhaps naively, she freely acknowledges) expected that physicians would use a computerized probabilistic model trained on large amounts of historical records and applied to her personal clinical data. Instead, she found that decisions were guided by the findings of clinical studies with sample sizes that represent only 3% of the 1.7 million patients diagnosed with cancer in the US each year. As Barzilay pointed out, imagine how well Amazon’s product recommendation engine would work if they discarded 97% of the data and used a biased subsample of only 3% of users.
Barzilay quickly learned that leveraging historical patient data was not as straightforward as she had hoped. Building accurate predictive models first requires extracting relevant data from patient records, and this is often a time- and labor-intensive process, especially since much of the most relevant data are stored in unstructured text. Barzilay then described some of her research on automating the extraction of relevant information from breast imaging pathology reports, including generating text-based explanations for model predictions and using transfer learning to adapt existing models to new categories. Barzilay used the last part of her talk to discuss her new line of research focused on directly analyzing mammograms and her goal of forecasting future malignancy from longitudinal data.

This talk offered two perspectives on how machine learners should seek to impact health care: (1) Target inefficiencies in the existing healthcare system, such as extraction of relevant information from clinical text. Breakthroughs in these areas could make the current system more effective and efficient. (2) Focus on solving problems for which no solutions currently exist, e.g., forecasting future malignancy. Both points of view have their merits, and we feel there is space for both kinds of research within our community.
Cynthia Rudin – Tools for Interpretable Machine Learning
Cynthia Rudin of Duke University continued the second session with a presentation on optimization methods for learning interpretable clinical prediction models. The investigation of interpretability remains a controversial topic in machine learning. Among doctors there is skepticism of so-called “black box” models and an affinity for simpler models. Classic clinical scoring systems consist of a linear sum of coefficients, similar to a regression, but where coefficients are restricted to be integers. This makes such scores easier to memorize and compute by hand. Rudin aims to develop more accurate and reliable scores using principled machine learning and reducing the reliance upon heuristics. Her proposed approach (described here and here) formulates the problem of learning a clinical score as a mixed integer linear program (MILP) with constraints. Rudin has used this approach to develop new scores for Attention Deficit Hyperactive Disorder (ADHD) and sleep apnea, among other diseases.
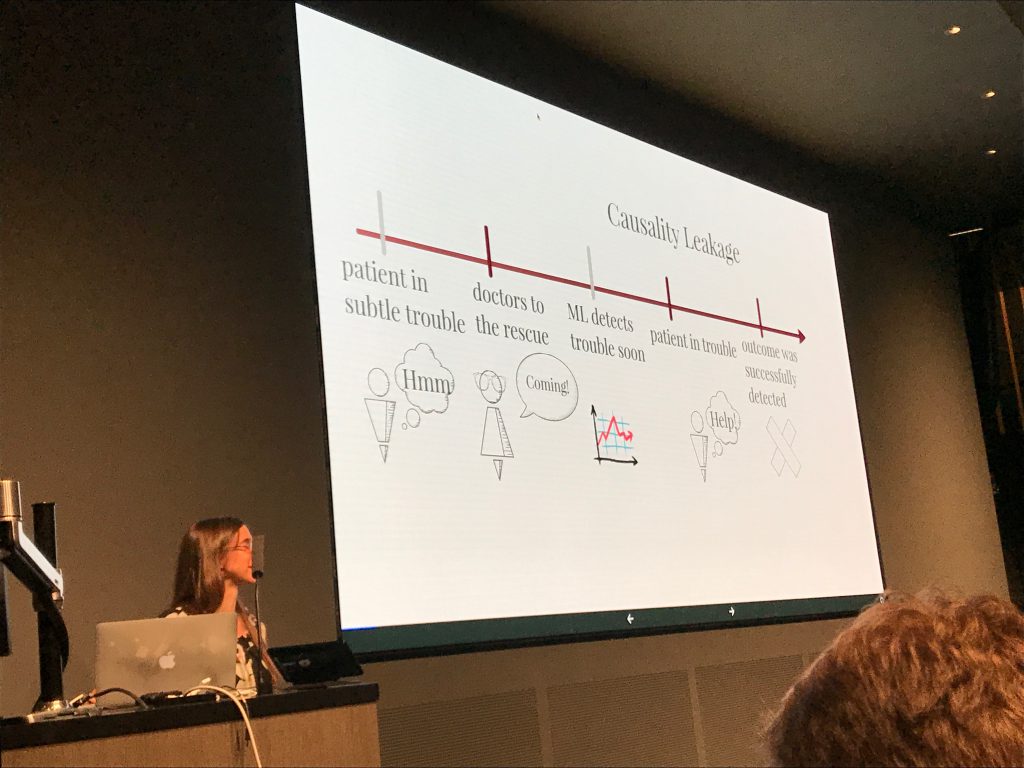
For us, the most interesting parts of the talk were the provocative statements that Rudin made about model interpretability and the debate that followed. She began with a claim that for many real-world problems, it is possible to learn an interpretable (simple) model that will be as accurate as less interpretable (complex) models. She connects this to Leo Breiman’s conjecture that there is often a “multiplicity of good models” (which he called the “Rashomon Effect” after the classic Japanese film). It worth noting that Breiman applied the Rashomon Effect to models within the same class of hypotheses, e.g., the many local optima for a given neural network architecture. It’s also worth noting that these methods only really make sense in domains where a tiny number of features confer most of the predictive power. In light of the widespread success, and adoption, of neural networks in complex domains, the idea that there is always a simple model that equals a more complex one in accuracy should raise doubts. In the QA session, Rudin acknowledged as much – these methods wouldn’t work well in perceptual domains (images, audio, signals) for example.
Rudin did leave us with this dank meme: “For this talk, an interpretable model is a function that fits on a powerpoint slide and can be computed without a calculator.” This definition addresses the notion of simulatability (compactness), but doesn’t address anumber of desiderata expressed by those concerned with interpretability in ML for healthcare. For example, this definition can still admit models that depend on spurious associations with no causal connection. Once nice feature of her definition, however, is that it is operational – it’s stated clearly enough that it can satisfied.
Saturday, August 19th
Dave Kale – Welcome Banter
(This section written by Zack for obvious reasons. To avoid any appearance of favoritism, it consists mostly of teasing Dave.)
In preparation for opening the second day, MLHC co-founder Dave Kale rambled for roughly ten minutes with the microphone on. As the room was empty, this caused no problems for the in-person conference attendees. However, anyone streaming remotely (see Facebook for archived stream), was treated to ten minutes of stream-of-consciousness about NIPS, beers, running, and speculation about whether or not Jenna was wrangling people to put down their coffees and come attend the morning session. While the conference venue was immaculate, the pristine environment was maintained by an ironclad no-coffee policy.
Once Jenna did in fact wrangle the crowd, Dave described the history of MUCMD to MLHC. Apparently, when Dave was a data scientist working for Randall Wetzell, director of anesthesia and critical care at CHLA, they had a grant for which they had promised to host a symposium. As the term of the grant drew to a close, they realized that they needed to haul ass and host a symposium quickly. Around this time, Dave had read a blog post (entitled “Regretting the Dead”) by ML luminary John Langford in which John reflected on the current state of clinical trials. Rather than force patients to endure bad treatments even after it’s known that the experimental arm works, Langford believed we could use the tools of reinforcement learning to design more adaptive experimental procedures. Randall exclaimed that these were the sorts of new ideas he wanted to hear at the symposium, and thus MLHC (then MUCMD) was born.
Matt Might – The algorithm for precision medicine
The first keynote on the second day came from Matt Might, whose background is neither machine learning nor healthcare but programming languages. A family health mystery led Matt to pivot, both personally and professionally into the field of precision medicine. His talk wove in and out of the story of his son Bertrand, who was born with a rare genetic movement disorder. Genetic analysis revealed that his son had two novel loss of function mutations in the NGLY-1 gene. When they discovered this condition, Bertrand was the first patient recognized to have this specific disorder and the gene itself was only described in five articles in the medical literature. The full story is described on a website dedicated to Bertrand and movement disorder.

Throughout the talk, Matt described the creative ways that he’s used social media, and collaborations with researchers in the life science to identify a community of patients with Bertrand’s condition (they’ve identified 59 to date). This work started with a viral blog post, and continued with Matt using his rising profile on Twitter and his search engine savvy to be discoverable by families Googling the relevant symptoms (e.g. “lack of tears”). He also described his attempts to try to treat Bertrand using only n-of-1 trials. These methods included some ad hoc safety testing, such as purchasing enzymes on Amazon and then taking them himself, and act he explained by “In my house, I am the FDA”.
Matt briefly commented on several areas where ML could impact health, but the biggest takeaway from his talk was that n-of-1 medicine can work. To that end, he concluded by describing some of the work he had done with the precision medicine initiative to try to help other patients with novel genetic disorders, like Bertrand’s, to discover effective treatments. Interestingly, Matt’s journey has led him to accept a position as Director of the Hugh Kaul Personalized Medicine Institute at the University of Alabama at Birmingham School of Medicine, an unusual position for someone whose academic career began with research on programming languages.
We should note that Matt’s talk continued an annual MUCMD/MLHC tradition of having at least one invited speaker present an unusual or outside perspective to the community.
Soojin Park – Machine Learning for Neurocritical Care
Concluding the morning session, Soojin Park, a specialist in neurocritical care spoke about opportunities to use machine learning to determine risk due to secondary injury from stroke. In a comprehensive talk she gave a window into this highly specialized area, highlighting some of the medial, computational, and administrative challenges. Many of her points hit home how high the stakes can be in the medical domain. For example, while working at MGH, she discovered that for many patients they received faster care for acute ischemic stroke if they went outside the hospital, called 911, and were picked up by an ambulance and admitted through the emergency room than if they had the event in an in-patient ward. Those currently in-patient received slower care mainly owing to process-oriented inefficiencies. The delay in care is so crucial because every minute without treatment following stroke corresponds to the loss of 2 million neurons and 14 billion synapses. According to some back-of-the-envelope math that’s roughly equal in some sense to 3.5 years of aging for every hour without treatment.
Apparently a large number of patients who get an initial stroke subsequently get a second stroke as a delayed effect of the first one. The secondary injuries can be difficult to recognize for a number of reasons. For one, the symptoms look like many other conditions that occur commonly in the hospital (symptoms include sedation, delirium). Moreover the monitoring that might help to recognize these events (CT-angiograms) cannot be done too frequently. Even a single CT-angiogram gives a non-trivial increase in the lifelong risk of cancer and cumulatively, with too many scans, a patient can suffer permanent hair loss among other symptoms. Her work aims to predict secondary brain injury from massive amounts of data collected from less invasive/risky sensors (paper).
Susan Murphy – Personalized Treatment in Real Time
Most current applications of machine learning to healthcare have the flavor of analytics, i.e., providing insights from historical data. However, for many, the ultimate hope is for ML agents to actually recommend beneficial interventions. There are some obvious obstacles to achieving this goal *interfering with the medical care of an actual patient could cause harm), and as a result, there’s very little research on this topic. One way around this is to act in a domain in which the set of actions are basically harmless. Susan Murphy of Harvard (previously U. Michigan) addressed one such domain – mobile fitness, where the actions are alerts or suggestions which prompt the user to, e.g., exercise.

Murphy’s talk led off with a snappy assessment of mobile health research: “This is the big conundrum in mobile health is not bothering people too much.” The research that she presented addressed patient activity. Each patient was equipped with a wearable mobile health sensor (like a FitBit) that measured step count, heart rate, and some crude measures of sleep quality. The device had the ability to prompt users, suggesting that they get up and walk around. Ultimately the goal was to coerce patients to be more active. The challenge is deciding when to make the suggestions. Murphy notes that when alerts come too frequently, users often either begin ignoring them or get pissed off and uninstall the app, a phenomenon known as “alarm fatigue” in clinical medicine. Murphy’s work found in an initial study that often these interventions can be more effective early in a study but that the effect can fade over time. For example, one experiment found that initially, patients responded to suggestions by taking 300-500 more steps per day but that by the end of the study this number fell to 60-70 steps.
One interesting observation was that suggestions were most effective when given at times of day where step counts were highly variable (as determined over previous 7 days). This makes sense, if a patient never takes any steps during a particular hour perhaps they are asleep or at work, or in the middle of a meal. The algorithmic approaches explored in the talk were simple formulations of the multi-armed bandit problem where the action is to make a suggestion (or not) and where the reward is the number of steps taken over the subsequent 30 minutes. It would seem that long-run effect on behavior (especially given alert fatigue observation) might depend on sequences of suggestions, and perhaps the next step algorithmically would be to think of this problem as a sequential decision process.
Collin Stultz – Risk Stratification in Cardiovascular Disease
The final invited talk on the second day came from Collin Stultz. His work primarily investigates the etiology of cardiovascular disease and while he holds an MD from Harvard Medical School, he also holds a PhD from MIT in biophysics and is currently a faculty member at MIT. He discussed methods for risk stratification. In short, the goal here is to recognize patients with a high risk of adverse outcome as early as possible. This information could be used by clinicians to make judgments about for which subset of patients the risk of death might be so high as to outweigh the risks of complications from invasive treatments. While he summarized the questions addressed in this line of research as (1) can we identify high risk patients? and (2) what interventions should we use?, the talk primarily addressed the former.
As input data for making risk predictions, he considered the standard risk score features, more comprehensive features based on patient history as available in electronic medical records, and morphological features based on analysis of electrocardiograms (ECGs). While it might be possible to feed raw ECG data into deep learning models, Stultz cautioned that the available data sets were considerably smaller than the hundreds of thousands of examples typically required when working with low-level perceptual data. Instead he and collaborators extracted two kinds of high-level features for the ECGs, length and slope of the ST segment. Using these two inputs at each time step to an RNN, they produced a model that improved upon the scores derived from health record data and conventional risk scores alone.
PANEL
The official conference ended with a panel-led discussion, moderated by Dave. Panelists included
- Daniel Golden, Director of Machine Learning, Arterys
- Susan Murphy, Professor of Statistics and of Computer Science, Harvard
- Binu Mathew, Vice President, Medical Intelligence and Analytics, Mercy
- Mark Homer, Director of Machine Learning and Informatics, Aetna
- Mark Sendak, Population Health Data Science Lead, Duke Institute for Health Innovation

The MLHC tradition is to pick a specific topic that spans a variety of concerns, not just clinical and computational but also social and practical. At MLHC 2016, the panel indulged in some science fiction, imagining the possible implications of a super-intelligent health AI and whether people would want their personal health micromanaged, even if it was guaranteed to be managed optimally. This year’s panel focused on a more mundane but far more immediate question: “What are the barriers that will prevent the work presented at MLHC 2017 from being deployed at the bedside in the future?”
The panelists discussed and debated a variety of issues, but focused in particular on several cultural gaps. For one, the panelists agreed that CS researchers cannot simply publish their cool new methods and then trust that the clinical community will adopt and deploy them. Most hospital systems are ill-equipped to do this alone and so CS researchers need to engage in the hard work of translation. This may require changing incentives within CS, where researchers are often judged primarily on their paper output (and sometimes their social media footprint!). Sendak indicated that there are several medical centers, such as Duke, that are prepared to engage with CS researchers who are committed to seeing their work deployed and tested in a real clinical environment.
The panel did not let clinicians off the hook! They discussed that because of the difficulties of deployment and integration with commercial EHRs, clinicians and hospital administrators often prefer to use simpler models, even when more accurate advanced models are available. While understandable and pragmatic, this attitude belies the community’s stated goal of building a “learning healthcare system.” The panelists argued that this may require a significant outreach and educational effort by the MLHC community. Mathew and Golden also suggested that CS folks need to communicate clearly to clinicians how new tools can impact their daily lives and help them do their jobs more efficiently.
THE PAPERS
If we went into similar depth about each paper accepted for publication, writing or reading this post would consume the fourth quarter of 2017. Instead, we provide links below to the accepted papers (which will be archived in a forthcoming issue of JMLR) and provide only a high level summary of the this year’s trends. The conference hosted 270 participants (at capacity) and received 66 submissions of which 26 survived the peer-review process. Among the 66 accepted papers at least two were first-authored by undergraduate students. In addition to the machine learning papers, the call for papers included a track for clinical abstracts and software demos. Among this latter category, six abstracts were accepted for poster and oral presentation. In its second year as a publishing conference, MLHC was chaired by its five founders, who oversaw a review process that included ten meta-reviewers and 91 total reviewers.
The impact of deep learning on the field was apparent with at least ten among the 26 accepted papers using deep learning techniques as their primary methodological contributions. Among the remaining papers, another three used neural networks in experimental comparisons. As mentioned in our discussion on the Arterys talk, surprisingly zero accepted papers used radiology data. While it’s possible that this indicates the field’s maturity and focus on translation, it’s also possible that this reflects the fact that only a small number of start-ups with hospital buy-in and bankrolling have sufficiently large datasets that researchers operating in public can’t compete using public data and benchmarks.
Of the accepted papers, nine addressed time series data. This is not surprising, owing to the natural temporal flavor of clinical data – patient care produces a stream of information that is observed in sequence and poses a number of decision problems which must be addressed in sequence. While this has always been the case, the uptick in papers on time series data may owe largely to the increasing prominence of recurrent neural networks, powerful tools which are capable of mining patterns in sequences. At least four papers addressed survival analysis and only four papers used the public benchmark MIMIC dataset. However, during the conference, we learned from one of the MIMIC creators that a newer, larger public dataset is now available on the physionet website. So we expect to see more work on public data at future events.
Among the grab-bag of more eclectic papers, one used computer vision (CV) for hand hygiene, another used CV for surgical skill estimation. One tried to quantify mental health based on internet postings. Most frighteningly, Alistair Johnson, one of the MIMIC curators, contributed a paper about reproducibility of studies using MIMIC data. He found that for many of the paper published with MIMIC data, even reproducing the cohort (subset of patients included in the study) was next to impossible, let alone reproducing the results. Many of the papers referencing MIMIC come from medical informatics venues which, in our experience, have very low technical bars. Still the finding should worry anyone working in this area and should motivate the community to adopt a practice of releasing code for both modeling and data processing, even when working with proprietary data.
Kvetching Session
Following the conference was a kvetching session in which the most hard-core attendees stayed late to offer constructive criticism for improving the conference next year. Overall the biggest challenges discussed were:
- How to bridge the culture gap among reviewers (papers typically have a clinical reviewer and a machine learning reviewer).
- While the event went off smoothly and was mostly successful, the demography made a hard shift towards ML (not enough clinicians attended). The consensus is that this happened in part because we didn’t host the conference at a hospital this year. Among the offered solutions are increasing outreach, always including a clinical organizer in the host city, and getting the necessary certification/registration so that clinicians can receive continuing education credits for attending.
Organizers
The organizing committee of five included three young computer science (CS) faculty (Finale Doshi-Velez of Harvard, Jenna Wiens of U. Michigan, and Byron Wallace of Northeastern), one CS PhD candidate (Dave Kale of the USC Information Sciences Institute, one of the co-authors of this post), and one clinician (Jim Fackler of Johns Hopkins). With the support of Carla E. Brodley, the Dean of the College of Computer and Information Science at NEU, the meeting was hosted at the brand new Interdisciplinary Science and Engineering Complex on the NEU campus.
SPONSORS
Several companies sponsored MLHC 2017, namely Google DeepMind, Aetna, Amazon, Microsoft Research, Cerner, Skymind, and the Laura P. and Leland K. Whittier Virtual Pediatric Intensive Care Unit (VPICU). Among the seven sponsors, four are tech companies – and all four sponsored MLHC in 2016 as well (VPICU supported MUCMD since 2011). The participation of Aetna and Cerner suggest that payors and vendors, perhaps taking notice of tech’s industry in health, are now competing for mindshare and talent.
What’s Next?
Machine learning for healthcare 2018 will be hosted at Stanford university. The call for papers is not out yet, but given history, the deadline should be around early April.
Thanks so much for this Zach, Dave!
Thanks a lot, Zach and Dave! As a budding ML researcher in healthcare, I found this very useful.